RStudio via OnDemand¶
Logging in¶
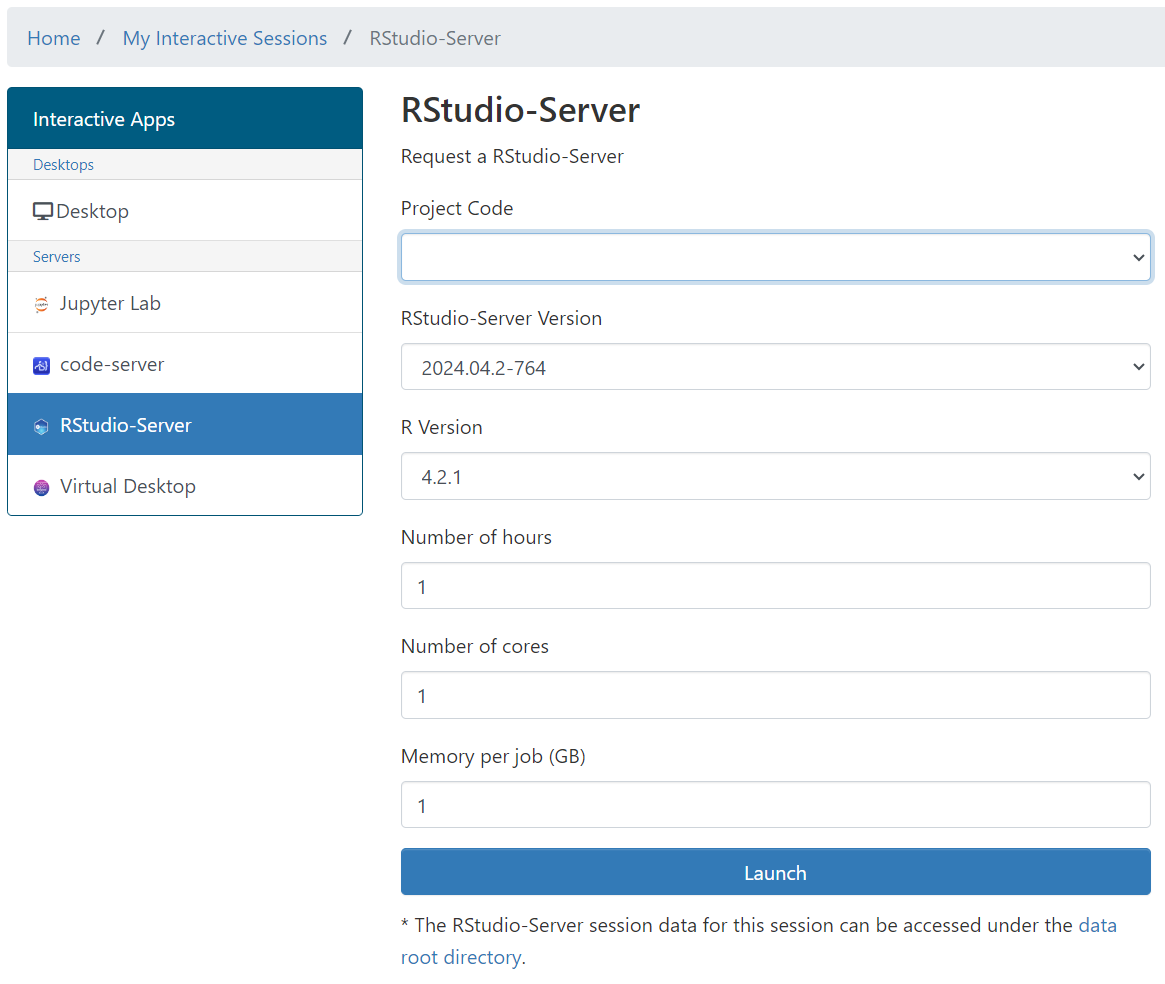
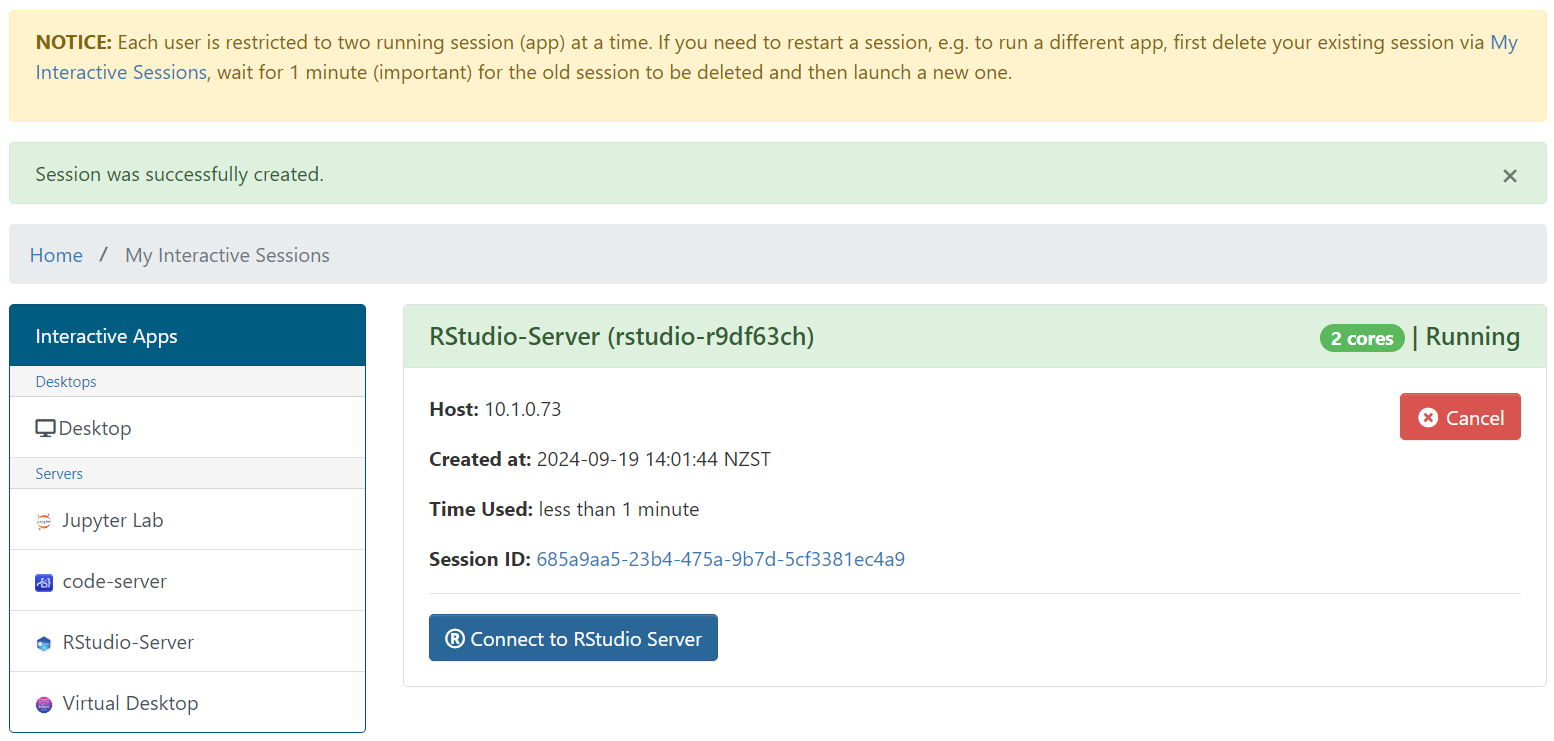
Settings¶
Recommendation to set Save Workspace to Never to avoid saving large files to the workspace. This can be done by going to Tools -> Global Options -> General and setting the Save workspace to .RData on exit to Never. This will prevent the workspace from being unable to load due to not enough memory in the selected session.
Bugs¶
Plots not showing¶
The current R modules on NeSI OnDemand do not support the default graphics device due to a missing depedency, cairo. There is a one off fix for this by changing the backend graphics device from Default to AGG (Anti-Grain Geometry) in the RStudio settings.
This can be done by going to Tools -> Global Options -> Graphics and switch Default to AGG. This will allow the plots to be displayed in the RStudio interface. You do not need to restart the RStudio session for this to take effect.
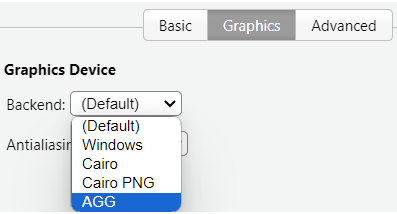
Modules from 4.4 onwards will have this issue fixed.
Libraries not showing¶
There is a bug with the R-Geo and R-bundle-Biocondutor libraries not showing up in the RStudio interface. This is a known issue and is being worked on. There are two workarounds for this issue:
- Manually add the library to
.libPaths()in the R console as shown below:
myPaths <- .libPaths()
myPaths <- c(myPaths, "/opt/nesi/CS400_centos7_bdw/R-Geo/4.3.2-foss-2023a")
# reorder paths
myPaths <- c(myPaths[1], myPaths[3], myPaths[2])
# reasign the library paths
.libPaths(myPaths)
# confirm the library paths
.libPaths()
[1] "/nesi/home/$USER/R/foss-2023a/4.3"
[2] "/opt/nesi/CS400_centos7_bdw/R-Geo/4.3.2-foss-2023a"
[3] "/opt/nesi/CS400_centos7_bdw/R/4.3.2-foss-2023a/lib64/R/library"
.Rprofile file in your home directory. This will automatically add the library path to the R console when it starts up. Copy and Paste the following lines to the file:
# CHECK LIBRARY PATHS
myPaths <- .libPaths()
newPaths <- c("/opt/nesi/CS400_centos7_bdw/R-Geo/4.3.1-gimkl-2022a",
"/opt/nesi/CS400_centos7_bdw/R-bundle-Bioconductor/3.17-gimkl-2022a-R-4.3.1>
# join the two lists
myPaths <- c(myPaths, newPaths)
# reassign the library paths
.libPaths(myPaths)